Burrows-Wheeler, Coronavirus, & Bayes
A Bioinformatics Pipeline to Detect & Understand COVID Mutants
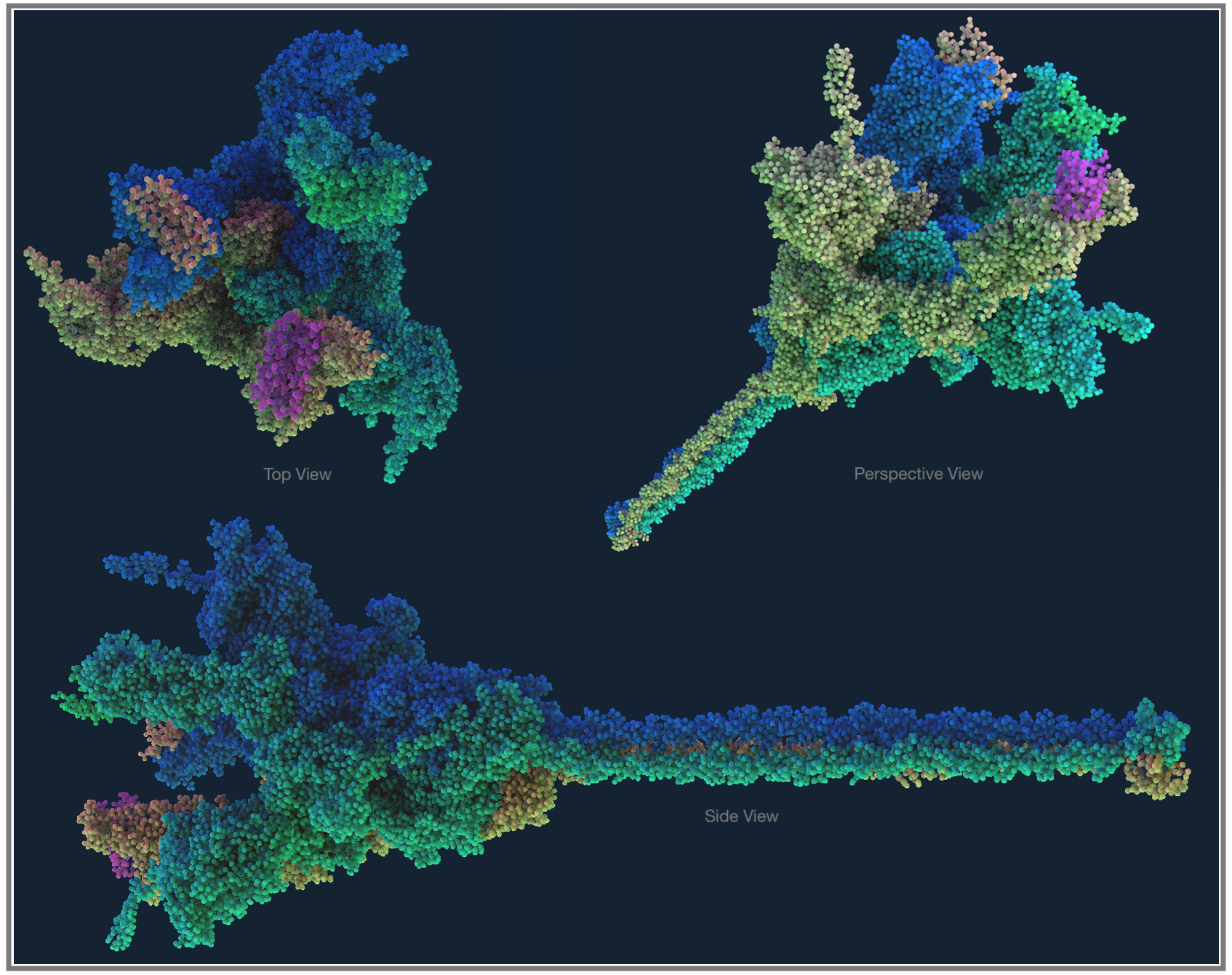
Fig 1. Molecular Structure with UK Mutant SARS-CoV-2 Docked to Human Receptor
As the pandemic runs its course, tracking genetic mutations has gained mainstream coverage. Officially, in the line of work we call bioinformatics, finding and understanding mutations is known as Variant Calling. In the same way every person is different from others (except twins), an individual virus can have variations in their genome; when a lot of these genetic variations accumulate between the virus we detected originally and what we find months later in a faraway town, we can say that we have a new strain of the virus. The Coronavirus which we’re now calling SARS-CoV-2 was first detected in Wuhan中国, so the genetic sequencing data from here is used as the base reference genome, and we measure mutations for the virus detected everywhere else against this; a coronavirus genome from a few towns away might only have a couple letters of the genome different from the Wuhan data, while something that’s travelled to the other side of the world after being transmitted through many people, climates, and even treatments, might now have acquired many more genetic variants compared to the original data.
Fig 2. Retrieving raw COVID data from SRA & creating alignment maps
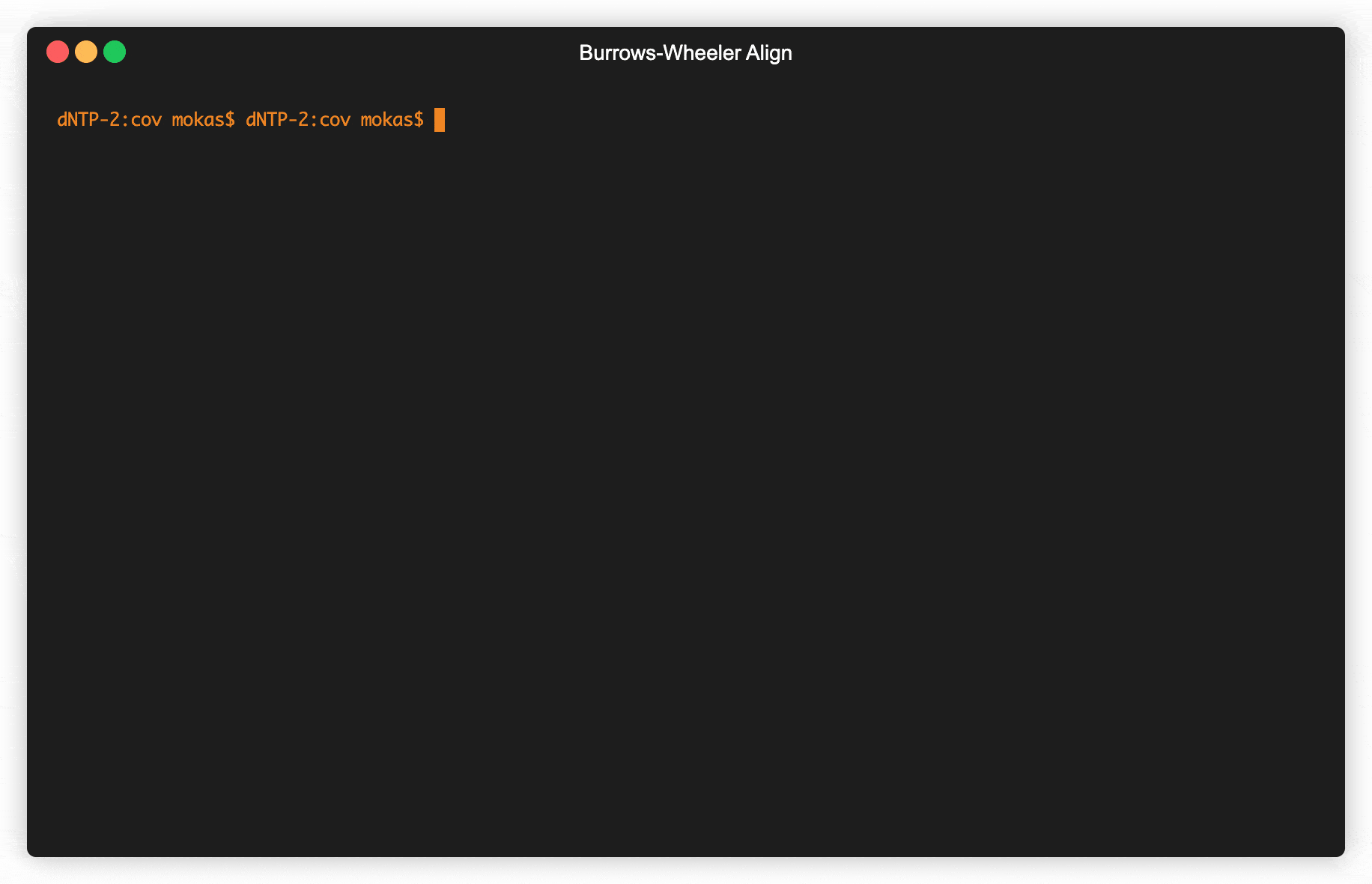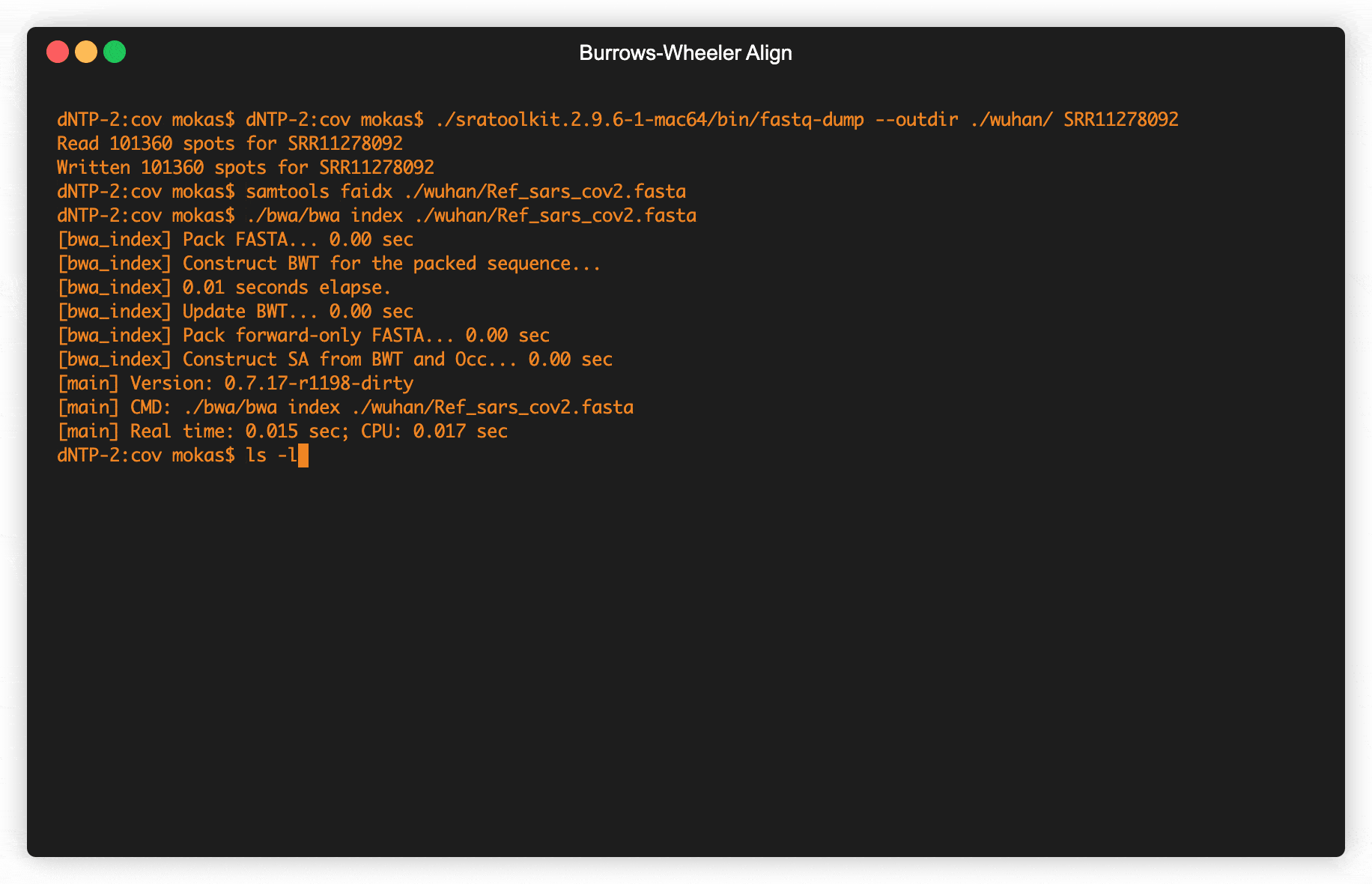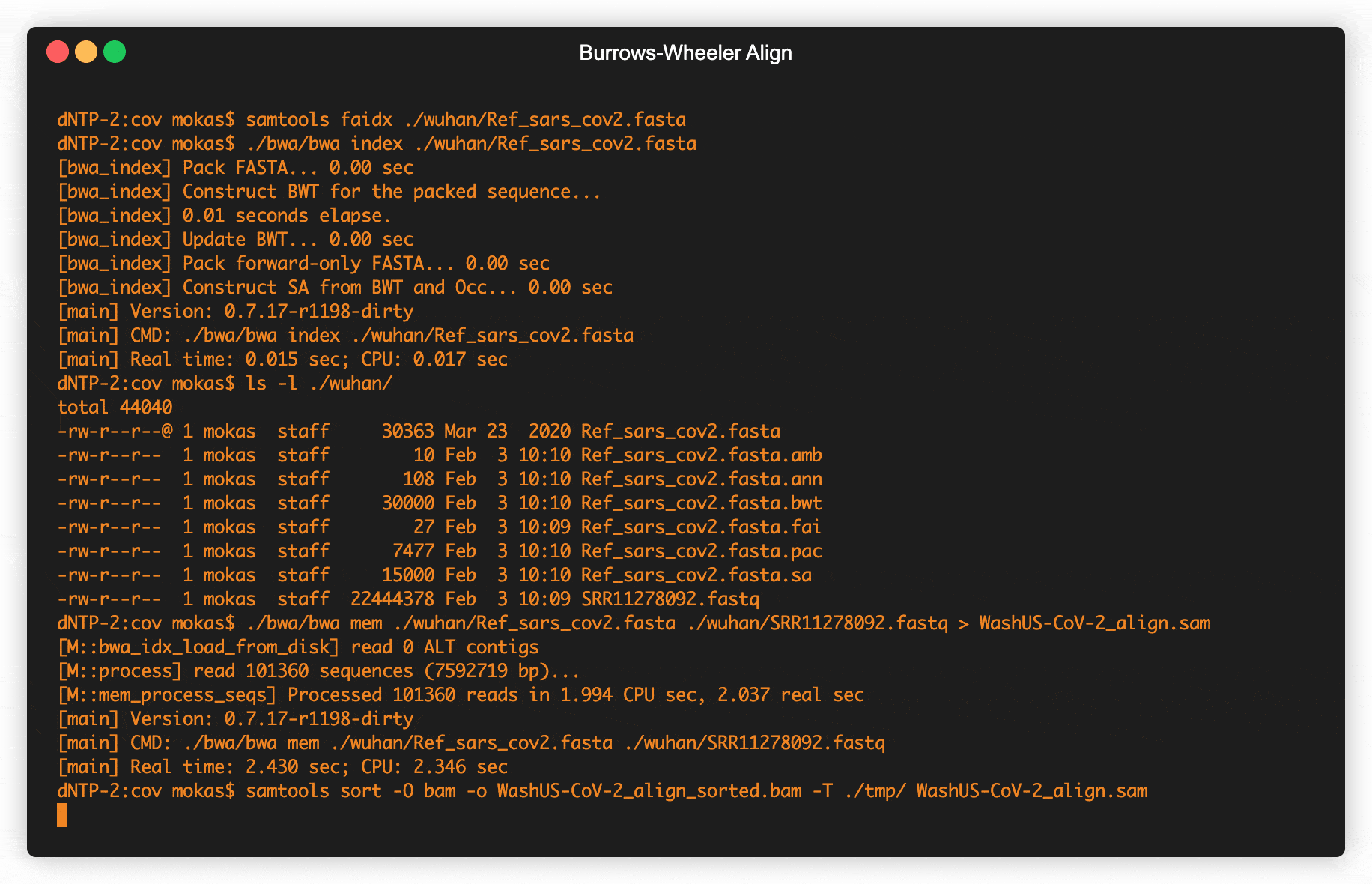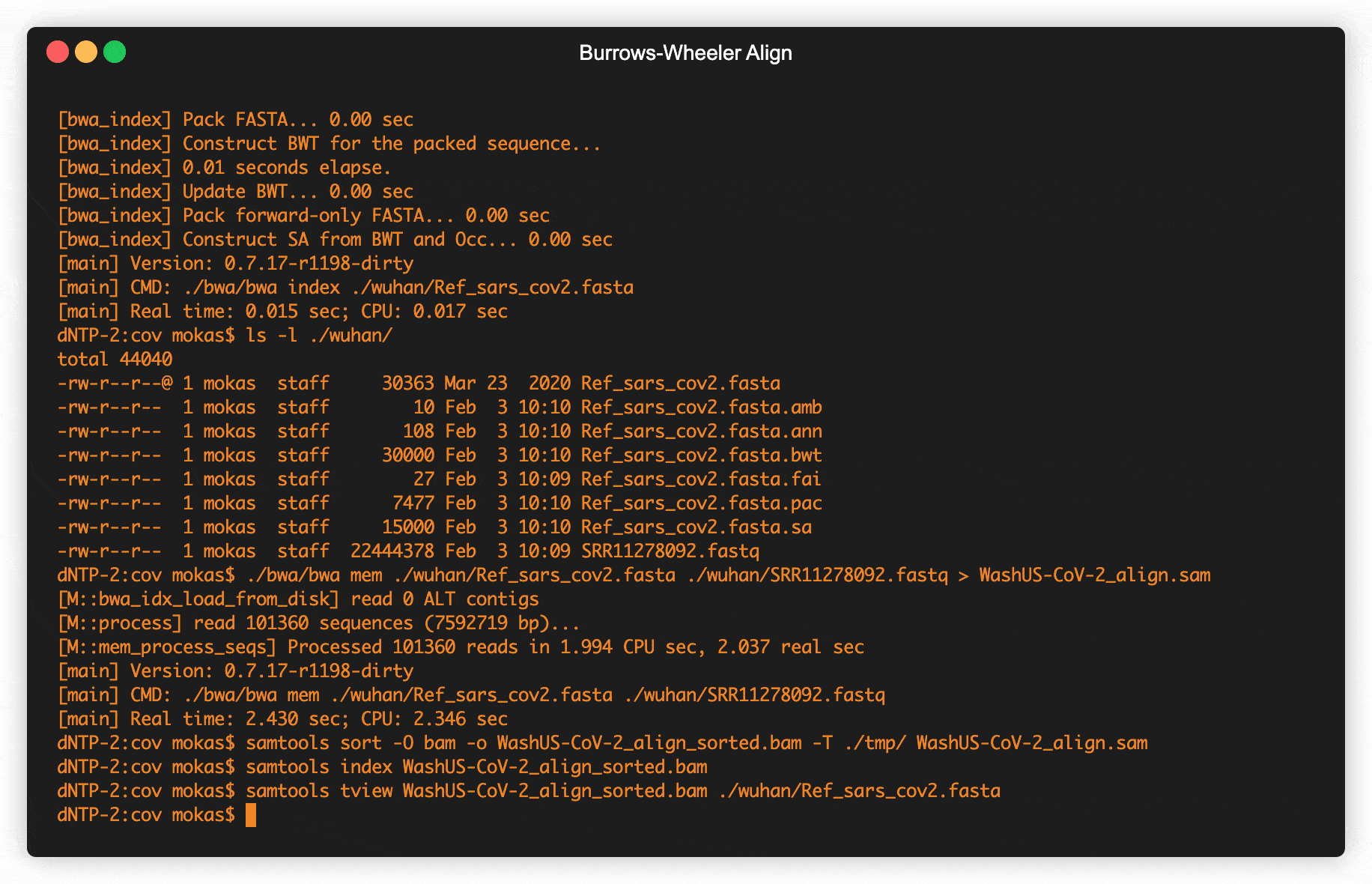 Data from viruses sequenced all across the world are uploaded into one of several databases (Table 1), this data is generated directly from machines by Illumina, Oxford Nanopore, or Life Technologies. When turning the molecules in genetic code into digital files these machines create strings that can be a few hundred letters in length to several thousand, but never the entire genome of the virus in a single string. A standardized file format called fastq holds all the strings from a single sample, including some quality data from the machine doing the sequencing. These strings are in no particular order, and first need to be mapped together to get the complete genome. Pretty much the standard way to create these genetic maps uses a technique invented in Palo Alto back in the early 90s by Michael Burrows and David Wheeler. The Burrows–Wheeler Transform was mostly just used in data compression, until around 2010 when computational biologists began to use it to align genomes together.
Data from viruses sequenced all across the world are uploaded into one of several databases (Table 1), this data is generated directly from machines by Illumina, Oxford Nanopore, or Life Technologies. When turning the molecules in genetic code into digital files these machines create strings that can be a few hundred letters in length to several thousand, but never the entire genome of the virus in a single string. A standardized file format called fastq holds all the strings from a single sample, including some quality data from the machine doing the sequencing. These strings are in no particular order, and first need to be mapped together to get the complete genome. Pretty much the standard way to create these genetic maps uses a technique invented in Palo Alto back in the early 90s by Michael Burrows and David Wheeler. The Burrows–Wheeler Transform was mostly just used in data compression, until around 2010 when computational biologists began to use it to align genomes together.
Nucleotide analog to clog Viral RNA Polyermase machinery in nCoV-2019
Remdesivir is for sale as of today, at $3,120 USD for the treatment of our current pandemic. Gilead Sciences has been devolping this Nucleotide analog for over 10 years. Nucleotide analogs look like one of the letters that DNA & RNA are made up of (this one looks like the letter “A”). But it isn’t that letter, it just looks like it, it’s analogous to it. So molecular machines that interact with these letters, nucleotides, accidentally end up using the analog instead and it messes things up.
Using SARS-CoV-2 Reference Genome to Align Outbreak Sequencing Data
In this post we use the reference genome established by the CDC: Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. To begin we need to grab just the FASTA data and save it as a file under something like nCoVref.fasta
