Using SARS-CoV-2 Reference Genome to Align Outbreak Sequencing Data
In this post we use the reference genome established by the CDC: Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. To begin we need to grab just the FASTA data and save it as a file under something like nCoVref.fasta
Next we will use samtools to index this reference file.
git clone https://github.com/samtools/samtools.git
cd samtools
./configure
make
make install
./samtools faidx nCoVref.fasta
This creates our nCoVref.fasta.fai index file, which will be useful later. To perform our actual alignment we are going to use the Burrow-Wheeler Aligner for short-reads, BWA.
git clone https://github.com/lh3/bwa.git
cd bwa
make
./bwa index nCoVref.fasta
BWA index created some additional files, like .bwt, .pac, .ann, and others. Now our reference genome is ready to align reads from sequencing data of patients from outbreaks. We are going to use isolate raw read files from the Washington SARS-CoV-2 outbreak uploaded to the Sequence Read Archive.To retrieve this data we need to use the SRA Toolkit, whose compiled binaries/install scripts can be found Here. Once installed we grab our reads:
./fastq-dump --outdir /path SRR11278092
We should now have a SRR11278092.fastq file. Which can be used with our previously prepared nCoVref file.
./bwa mem nCoVref.fasta SRR11278092.fastq > SARS-CoV-2_align.sam
./samtools view -bS SRR11278092.sam | samtools sort - SARS-CoV-2_align_sorted
Our output from the last few steps should be a SARS-CoV-2_align_sorted.bam file. This file also needs to be indexed so that we can navigate it quickly:
./samtools index SARS-CoV-2_align_sorted.bam
Samtools can be used to view the alignment maps just created:
./samtools tview SARS-CoV-2_align_sorted.bam nCoVref.fasta
Tview can be very slow and difficult to use for anything other than a quick look because terminal has a difficult time rendering all the charecters and colors. Below are two external tools we can use, including one that I work on.
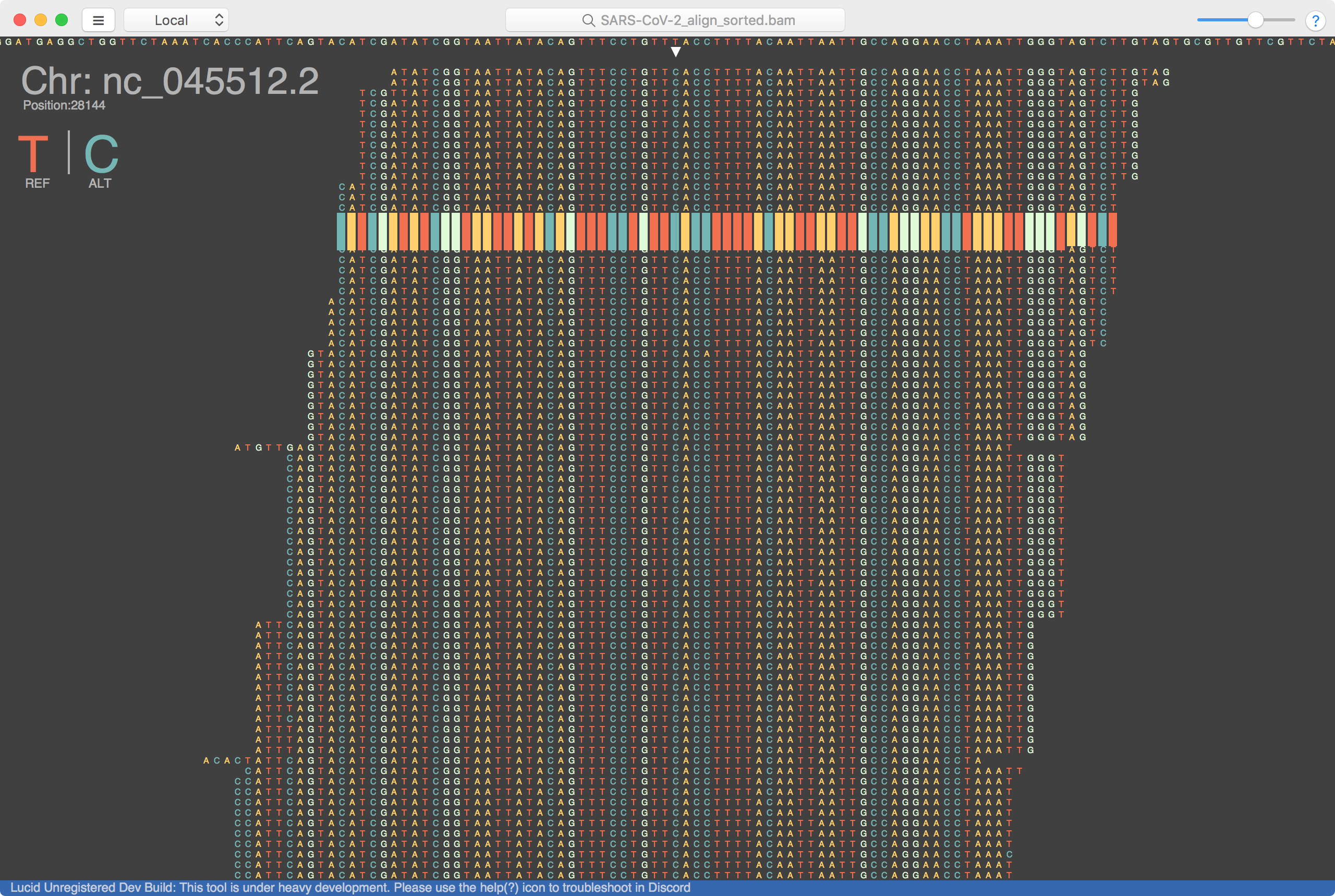
Lucid is much faster than using the terminal, and doesn’t have any problems rendering the data since it’s written in C#. However, The Integrative Genomics Viewer (IGV) is really the academic standard when it comes navigating sequence alignments. Although it’s not as quick or pretty, it has many more esoteric features which can be helpful to niche projects.
Using these viewers is only the first use of the reference & alignments we prepared. It’s helpful in looking directly at specific mutations, and genes with a human eye. But at scale we will need to use variant callers, and maybe go back and forth between an automated process and some manual inspection. We can already see some mutations between the reference genome from the Wuhan samples to the alignments from Washington. Will try to post about scaling up & variant calling soon.
Happy Virus Hunting!
